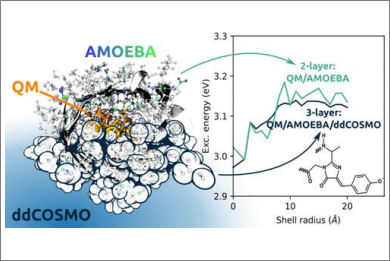
We present the implementation of a fully coupled polarizable QM/MM/continuum model based on the AMOEBA polarizable force field and the domain decomposition implementation of the conductor-like screening model. Energies, response properties, and analytical gradients with respect to both QM and MM nuclear positions are available, and a generic, atomistic cavity can be employed. The model is linear scaling in memory requirements and computational cost with respect to the number of classical atoms and is therefore suited to model large, complex systems. Using three variants of the green-fluorescent protein, we investigate the overall computational cost of such calculations and the effect of the continuum model on the convergence of the computed properties with respect to the size of the embedding. We also demonstrate the fundamental role of polarization effects by comparing polarizable and nonpolarizable embeddings to fully QM ones.
Nottoli, M.; Nifosì, R.; Mennucci, B. & Lipparini, F.
J. Chem. Theory Comput. 17, 5661–5672 (2021) https://doi.org/10.1021/acs.jctc.1c00555